Abstract
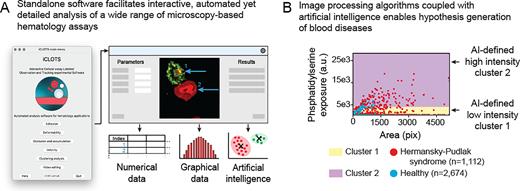
While significant effort has been devoted to developing novel in vitro microscopy-based hematology assays, a clear need exists for new software-based tools to analyze the data-rich images and videos obtained thereof. Whereas traditional manual analysis of cellular microscopy data is time-consuming and error-prone, advanced computational methods require substantial coding expertise. Popular general-purpose bioimage analysis software including ImageJ, Ilastik, and CellProfiler have robust image processing and visualization algorithms, however, they cannot adapt to hematology-based tools, e,g. time course-based experiments utilizing commercially available flow chambers or novel microfluidic devices, without significant additional calculations or coding/scripting. Accordingly, we developed a user-adaptable suite of tools packaged into an open-source, standalone software, "iCLOTS” (interactive cellular assay labeled observation and tracking software). iCLOTS comprises pre-defined workflows based upon well-validated image processing and artificial intelligence (AI) algorithms for specific use with blood cell-based assays. Designed as a post-processing software, iCLOTS is suitable for previously-collected data or data collected with iCLOTS capabilities in mind. iCLOTS blood cell adhesion, cell deformability, fluid flow-based velocity experiments, and microfluidic-centric applications generate automatically labeled quantitative numerical and graphical data from user-provided files. To assist researchers in interpreting potentially large datasets comprising multiple metrics which describe up to thousands of cells, our software also capitalizes on advances in the burgeoning field of machine learning (ML), a subset of AI. ML clustering algorithms mathematically characterize previously imperceptible data groupings, enabling generation of novel hypotheses with clear implications for digital pathology, personalized medicine, and establishing clinical diagnostic reference ranges (Figure 1A).
We demonstrate iCLOTS’ capabilities as a research-enabling platform by reporting new discoveries related to inherited platelet disorders, sepsis, and sickle cell disease. For example, we investigate Hermansky-Pudlak syndrome (HPS), an hereditary platelet disorder that results in platelet dysfunction with prolonged bleeding. Analysis of fluorescence microscopy images reveals increased spreading and phosphatidylserine (PS) exposure via annexin V labeling in platelets obtained from patients with HPS as compared to healthy patient controls on collagen-coated surfaces (n=2,674 and n=1,112 platelets, respectively). PS exposure on the surface of activated platelets confers a procoagulant surface necessary for hemostasis (Kay and Grinstein, Lipid-mediated Protein Signaling, 2013). ML clustering using k-means algorithms automatically separates all single-platelet data points into mathematically optimized clusters describing low and high PS-exposure groupings, in a method akin to flow cytometry with objective boundaries. We find a significantly greater proportion of HPS platelets in the high-PS-exposure cluster as compared to healthy control platelets (*p < 0.001 via Chi-squared test, Figure 1B).
iCLOTS is presented to the hematology community as an interactive, standalone software that enables all hematologists to discover a new level of automated analyses for their microscopy-based data. Researchers and clinicians can explore questions about both fundamental cell processes and patho/physiology via measures of biological functionality, cell mechanical properties, properties of cell suspensions, and/or microchannel behavior. Designed to work with all blood cell types and a range of experimental systems, iCLOTS produces detailed numerical results describing a range of cell characteristics and behaviors which may be further assessed by ML algorithms capable of detection of healthy/clinical sample dichotomies and/or significant subpopulations of blood cells. Built by hematologists, for hematologists, iCLOTS balances versatility and automation with plug-and-play capabilities, requiring no computational expertise, thereby enabling a new level of single-cell resolution data analysis free for the entire field of hematology to use/download.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal